High Performance Computing
High performance computing (HPC) encompasses a broad range of advanced computing capabilities, from single GPUs to the world’s largest supercomputers. From enabling simulation of intricate biological processes to providing real-time feedback in surgical settings and advancing machine learning applications—to name a few—HPC helps researchers explore new frontiers in medicine.
At the Duke Center for Computational and Digital Health Innovation, HPC serves as a foundational technology for enabling groundbreaking research and discoveries in medicine. Through state-of-the-art supercomputing resources, we unlock the potential to solve complex biological challenges and drive transformative discoveries.
As technology progresses, the Center continues to leverage HPC to find, track, and treat medical conditions, setting the stage for innovative and effective healthcare solutions.
What Is High Performance Computing?
High performance computing refers to the use of advanced computational systems to process vast amounts of data and run high-fidelity simulations at unparalleled speeds and scales.
Researchers at the Center use onsite clusters with state-of-the-art GPUs, cloud computing platforms, and leadership-class supercomputers to advance their research.
What Are the Advantages of High Performance Computing?
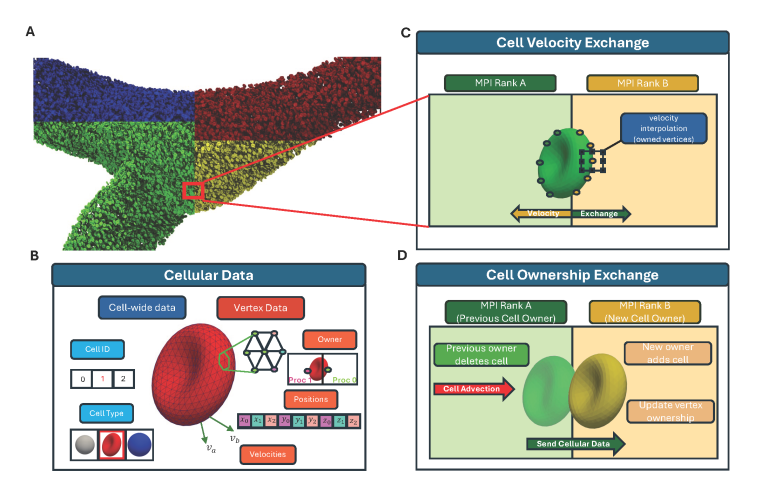
HPC allows our researchers to overcome significant engineering challenges, tackle larger spatial domains, and extend the temporal scope of their studies. By reducing overall runtime, HPC enables real-time feedback and accelerates the pace of medical research.
Our researchers use HPC to support a broad spectrum of healthcare advancements, including:
- AI training: We train advanced artificial intelligence models on large datasets to uncover patterns, detect diseases, and predict health outcomes with precision.
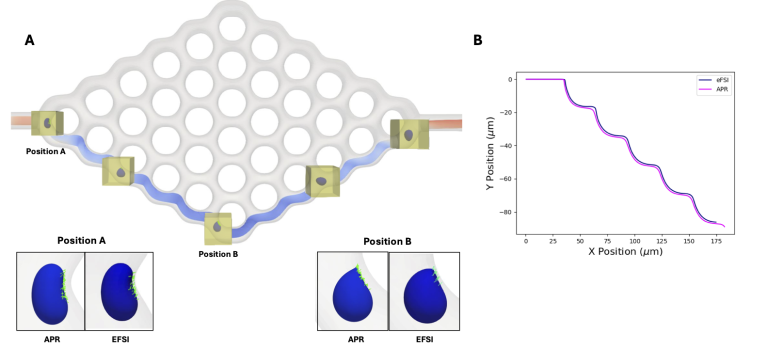
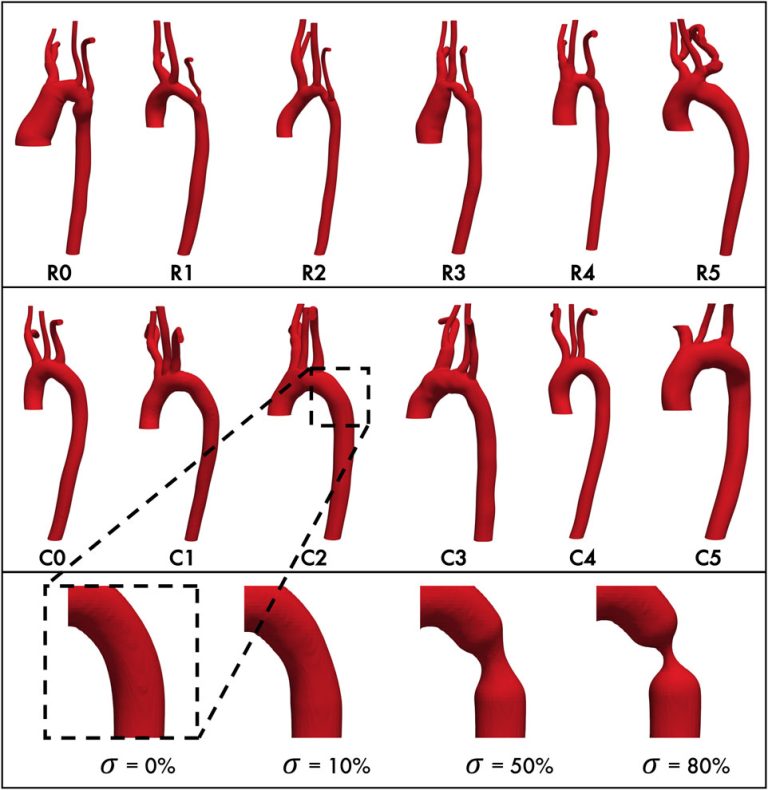
- Computational modeling: By using HPC, we can simulate longer temporal domains and larger spatial domains; produce higher resolution data, larger step times, and capture larger areas; and reduce time to solution.
- Data analytics: We process and analyze time-series data from wearable sensors and clinical records to deliver actionable insights in real time.
- Cloud computing integration: We use scalable cloud platforms to enhance computational efficiency and ensure accessibility across interdisciplinary teams.
Through the skills of our researchers, these powerful capabilities enable us to address medical challenges that were previously considered impossible to solve.
How Do We Use High Performance Computing?
High performance computing is a transformative tool at the Center for Computational and Digital Health Innovation. Our researchers are driving significant advancements in medical research and clinical practice through it. A few examples follow.
Real-Time Computing for Neural Speech Prostheses
The collaborative efforts between the Viventi and Cogan Labs focus on developing advanced neural interfaces and real-time computing solutions to restore communication for patients with neurodegenerative diseases. These patients often lose the ability to speak, severely impacting their quality of life. The team addresses this challenge by utilizing high-resolution, micro-electrocorticographic (µECoG) neural recordings to capture detailed brain activity during speech production. This novel approach provides neural signals with significantly higher spatial resolution and signal-to-noise ratio compared to standard invasive recordings, resulting in nearly double the decoding accuracy.
The use of advanced computing techniques, including non-linear decoding models, leverages the rich spatio-temporal structure of these neural signals to enhance speech decoding. The real-time aspect of this research is crucial, as it enables the development of neural speech prostheses that can operate in real-time, allowing for immediate feedback and communication. Additionally, the project addresses the unique challenges of deploying these systems in environments without reliable Wi-Fi connectivity, ensuring that the technology can be used effectively in various settings. The interdisciplinary collaboration between the medical and engineering schools at Duke enhances the development and application of these cutting-edge technologies, providing new hope for patients with speech impairments.
Real-Time Computing for Neural Speech Prostheses
The collaborative efforts between the Viventi and Cogan Labs focus on developing advanced neural interfaces and real-time computing solutions to restore communication for patients with neurodegenerative diseases. These patients often lose the ability to speak, severely impacting their quality of life.
The team addresses this challenge by utilizing high-resolution, micro-electrocorticographic (µECoG) neural recordings to capture detailed brain activity during speech production. This novel approach provides neural signals with significantly higher spatial resolution and signal-to-noise ratio compared to standard invasive recordings, resulting in nearly double the decoding accuracy.
The use of advanced computing techniques, including non-linear decoding models, leverages the rich spatio-temporal structure of these neural signals to enhance speech decoding. The real-time aspect of this research is crucial, as it enables the development of neural speech prostheses that can operate in real-time, allowing for immediate feedback and communication. Additionally, the project addresses the unique challenges of deploying these systems in environments without reliable Wi-Fi connectivity, ensuring that the technology can be used effectively in various settings.
The interdisciplinary collaboration between the medical and engineering schools at Duke enhances the development and application of these cutting-edge technologies, providing new hope for patients with speech impairments.
Advancing Machine Learning in Biomedical Research
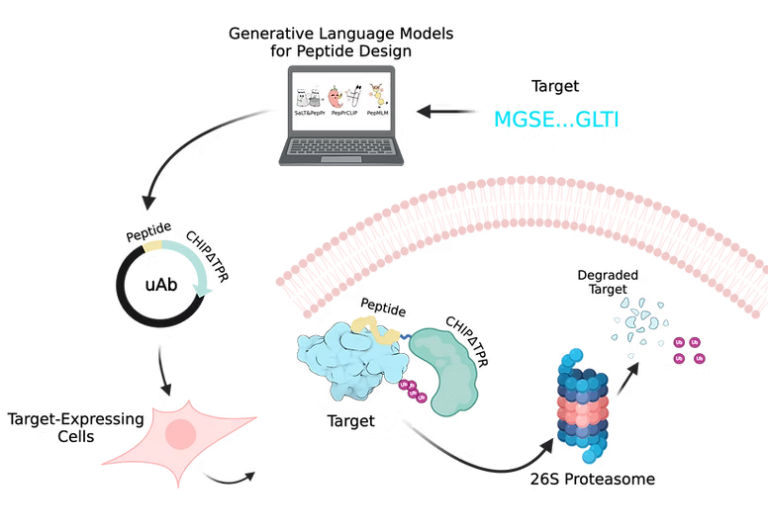
The Chatterjee Lab harnesses the power of HPC and GPUs to develop advanced machine-learning and AI models that support its protein design efforts. The lab focuses on three main areas:
- Programmable proteome editing
- Programmable genome editing
- Programmable cell engineering
By leveraging GPUs, the lab can efficiently train generative language models such as Cut&CLIP, SaLT&PepPr, PepPrCLIP, and PepMLM. These models help with the design of peptide-guided therapeutics, CRISPR-mediated genome editing tools, and protocols for differentiating ovarian cell types from pluripotent stem cells.
The computational power of GPUs accelerates the lab’s ability to process large datasets and complex algorithms, facilitating rapid advancements in targeted therapeutics and genetic engineering.